Force Multipliers
GW’s interdisciplinary research teams soar to new
heights.
By Kathleen Kocks
Clearly, interdisciplinary research is a force multiplier.
It joins diverse scientific disciplines in collaborative efforts
that multiply the effectiveness of research. The results are
often developments that otherwise would be impossible.
“Each discipline brings a different culture, ways of
thinking and analyzing problems,” explains one collaborator
involved in several interdisciplinary projects at GW, James
Hahn, professor and chair of the Department of Computer Science.
“By working together in an interdisciplinary environment,
not only do we learn about and appreciate the other disciplines,
but we learn to look at our own disciplines with a wider perspective.”
Many opportunities exist for interdisciplinary research within
GW’s numerous schools and institutes. Such research
also has a strong proponent in Chief Research Officer Elliot
Hirshman.
“Contemporary research universities must discover the
knowledge and create the technologies necessary for our society
to advance and prosper,” Hirshman says. “In this
context, researchers are attempting to ensure that we have
sufficient energy resources, a sustainable environment, homeland
security, a productive, competitive economy, and innovative
and affordable health care. Solving these complex problems
requires researchers from multiple disciplines to work together,
making interdisciplinary research central to the contemporary
research university.
“GW’s researchers have recognized the importance
of interdisciplinary collaboration and have formed multidisciplinary
teams including engineers, scientists, and clinical researchers,”
he continues. “This substantial grassroots movement
represents our greatest strength.

Eliot Hirshman
Jessica McConnell
|

|
“Working with the deans and the associate vice president
for health research, the central administration has two roles.
The first role is to support the extant teams on campus—providing
them with the financial resources and administrative support
they need to prepare competitive proposals and carry out their
scientific projects. The second role is to identify external
funding opportunities and to facilitate the formation of interdisciplinary
teams to respond to these opportunities.”
Attesting to Hirshman’s support is Patricia Berg, associate
professor of biochemistry and molecular biology, who is co-leading
an interdisciplinary project to develop a blood test to detect
breast cancer. “The existence of our project owes much
to Elliot Hirshman, who has the vision to understand the importance
of collaborative research,” Berg states. “He is
actively fostering interdisciplinary research throughout GW
and obtained the seed money we needed to give our project
a real boost forward.”
Berg’s project and three others featured here are excellent
examples of the power of interdisciplinary research at GW.
Breast Cancer Biosensor
Patricia Berg: Associate Professor,
Biochemistry and Molecular Biology
Robert Siegel: Professor, Medicine
Samuel Simmens: Interim Director, Biostatistics
Center Medical Center Unit
Akos Vertes: Professor, Chemistry,
Biochemistry and Molecular Biology
Mona Zaghloul: Professor, Engineering
and Applied Science
In 2003, GW Medical Center’s Berg led a team from four
institutions to discover that a gene she had studied for 16
years—BP1, for beta protein 1—is activated in
the tumors of 80 percent of women with breast cancer. Subsequently,
she discovered the same gene plays a role in 70 percent of
prostate cancer cases and in 63 percent of acute myeloid leukemia
cases. This discovery indicates that BP1 may figure prominently
in other types of human cancer.
Berg also discovered that the presence of BP1 increases as
breast cancer progresses, and it is activated in breast tumors
that no longer have a certain protein, called an estrogen
receptor. Furthermore, breast cancer patients lacking this
protein have a poor prognosis.
Today, Berg is building upon those discoveries and banking
on interdisciplinary research to develop a blood test to detect
the presence or lack of this protein and many others. The
project teams Berg’s expertise in biochemistry and molecular
biology with expertise in engineering, microelectro-mechanical
systems (MEMS), oncology, and biochemistry.
“The goal of our research is to develop a blood test
that uses a MEMS-based biosensor to detect BPI, as well as
other proteins that are released from breast tissue in the
earliest stages of cancer,” Berg says. “Such a
test would have three distinct benefits: extremely early detection
of new cancer cells, monitoring the progress of breast cancer
treatment, and early detection of recurring cancer. Today,
no blood test does this.”

|

This photo shows an ultra high performance liquid chromatograph,
which is used to separate proteins from complex biological
mixtures.
Jessica McConnell
|
Contributor of the engineering expertise and co-leader of
the project is Mona Zaghloul, professor of engineering and
applied science, who previously produced a sensor that was
deemed appropriate for this project. It was originally developed
to detect trace amounts of chemicals—something of interest
to government agencies fighting terrorism.
“We are also working with Professor Akos Vertes, who
is helping us with the chemistry we need to develop the biosensor,”
Berg says. “The plan is to coat the biosensor microchip
with gold, then attach to it various molecules, including
antibodies specific for the proteins we are trying to detect.”
“If a protein is present,” Berg explains, “it
will bind to its antibody and cause a change in the frequency
of the biosensor’s current. The biosensor will produce
an electronic readout showing the level of the various proteins
in the blood.”
The team is hoping to eventually attach antibodies for numerous
different proteins to the biosensor. However, the work is
beginning with antibodies to one well-known protein called
mammaglobin, which is detected in the blood of women who have
metastatic breast cancers. Robert Siegel, professor of medicine
and a hematologist/oncologist, will provide blood samples
from breast cancer patients.
“This will be our model system to determine if the
biosensor is able to detect the protein,” Berg explains.
“Two advantages of the biosensor are its small size
and high sensitivity, so we only need a very small amount
of blood to get results. The biosensor is also capable of
multiplexing, so it can simultaneously detect different proteins.
This will facilitate testing and also could help us further
understand the interaction among proteins—that is, how
one protein may work with or against another protein.”
The project began in October 2005. One of Zaghloul’s
students, Onur Tigli, is creating the gold microchips. Cynthia
Chatterjee and Lou Bivona, researchers in Berg’s lab,
have successfully attached the first antibody to the gold
surface.
The project is taking a building-block approach, starting
by using a purified protein with a purified antibody to see
if the sensor performs as anticipated. Next will come testing
of a mix of purified and normal materials, then finally testing
of patients’ blood. Samuel Simmens, interim director
of the Biostatistics Center Medical Center Unit, and another
member of the team, will perform statistical tests for significance
of the data after analysis of the blood samples.
“While this research project is only focusing on breast
cancer, successful development of our biosensor would have
a much larger applicability,” Berg concludes. “Scientists
could develop a biosensor for any condition in which one wanted
to detect a protein in the blood.”
Biomolecular Signaling Networks
Rahul Simha: Professor, Engineering
and Applied Science
Frank Turano: Associate Professor,
Biology
Chen Zeng: Associate Professor,
Physics
Plants, unlike most organisms, do not have the advantage
of mobility. They cannot, for example, move from shade to
sun, or from wet to dry. Nor can they move from an unhealthy
soil to a nutrient-rich soil. Plants react to stress through
a series of molecular events that signify changes are coming,
and the plants ultimately produce compounds to survive the
stress. How this information flows through the plant is the
focus of an interdisciplinary project joining biology, physics,
and computer science.
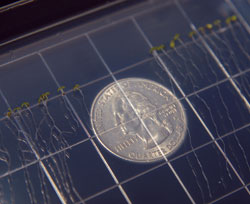
To better understand how plants react to stress, Turano’s
team uses the weed Arabidopsis to explore how information
flows through it. Quarter used to show scale.
Jessica McConnell
|

|
“We are trying to understand a biomolecular signaling
network,” says Frank Turano, associate professor of
biology. “How do plants change their metabolism to address
stress? It turns out that the information does not simply
flow in a straight line. The process operates more like a
huge hub-and-spoke network; most similar to a map of airline
routes, in which the airports represent hubs. And like the
airports in the analogy, some hubs are vital to the signaling
network, but others are not. We are trying to find out which
information hubs are vital.”
To conduct this research, plant biologists have two invaluable
tools. One is a research model described as the perfect “guinea
pig” of the plant world: the weed Arabidopsis.
“Scientists have sequenced its entire genome, so we
know all the genes,” Turano explains. “We also
have a molecular microchip that allows us to conduct experiments
to determine what happens to all the genes in the plant simultaneously.”
Turano’s team is giving plants different compounds
and recording the expression of all 27,000 genes to obtain
an accurate road map of how the signals move within the plant.
“As we piece information together, we are trying to
understand the topology of how the information is sent,”
Turano says. “Can we see which way the signals are flowing?
Are there information hubs? And if so, which hubs are important?”
Working with Turano are Chen Zeng, associate professor of
physics, and Rahul Simha, professor of engineering and applied
science. Together they oversee research activities in the
“Institute for Biomolecular Networks,” partly
funded by GW’s Research Enhancement Fund. The institute’s
objective is to develop algorithms and mathematical models
to simulate how the information flows and how each hub and
circuit performs. For the moment, the project’s team
is looking at small and fairly well-known molecular hubs and
circuits.
“Once we can simulate the simple hubs and circuits,
we can build up to developing models for the more complex
signaling events and create an information networking tool,”
Turano explains. “If we are able to mathematically model
and computationally simulate the operation of these hubs and
reliably predict metabolic behavior, then hopefully we can
expedite the way we do research. Rather than testing randomly,
we could use the model to simulate how the information flow
works, then go to the plant and see if the model accurately
predicts the outcome.
“Then we will find out what happens if we knock out
individual hubs. Will it kill the plant, or will it make the
plants bigger or smaller than normal? So far, we have been
able to knock out an important hub by turning off a gene and
studying how this affects the other signaling events.”
The practical applications for this research could be significant.
Turano ultimately wants to understand how plants respond to
changes in their environment, hoping to find ways to grow
plants more efficiently. This could greatly improve crop productivity,
while also decreasing the need for fertilizers and herbicides.
“Overall, however, the value is in understanding biological
systems in general. You cannot resolve problems if you do
not understand how something works,” Turano states.
“If we can understand how information flows in a cell,
model it, and simulate it, then we can modify and optimize
the plant itself.”
Protein Microscope
Eric Hoffman: Professor, Pediatrics,
Biochemistry, and Molecular Biology
Fatah Kashanchi: Professor, Biochemistry
and Molecular Biology
Mark Reeves: Professor, Physics
Akos Vertes: Professor, Chemistry,
Biochemistry and Molecular Biology
Proteins are the basic building blocks of all cells. They
also communicate signals to and from the body’s components.
They ensure good health but can cause disease when nature
goes awry. Because proteins are so vital, scientists are seeking
new ways to better understand them.
Development of a tool for such research is the goal of the
protein microscope project at GW’s Institute for Proteomics
Technology and Applications. Joining experts in physics, chemistry,
biochemistry, and medicine, this interdisciplinary project
began in 2004 and has support from the W.M. Keck Foundation—which
funds projects that are innovative and expected to have a
large impact—along with other foundations and government
agencies.

|

Shown is the sample stage of the scanning near-field
optical microscope. After combining it with the mass
spectrometer, it will become an integral part of the
protein microscope.
Jessica McConnell
|
“The protein microscope is being developed for the
science of proteomics: the study of proteins, their structures,
and their functions,” explains Professor of Physics
Mark Reeves, who is part of the team building the project’s
scanning near-field optical microscope. “Understanding
which proteins are involved in the body’s processes
and how they work is very important. In the traditional approach
to detecting proteins, tissue material is obtained, ground
up, and then observed for the presence or lack of proteins.
But this approach obscures information on precisely when and
where proteins are expressed, making it difficult to interpret
and understand the protein’s role.
“The protein microscope overcomes this limitation and
allows us to zero in on the exact spot in an individual cell
where the protein’s activity is occurring. It enables
us to measure which proteins are active and which are not,
as well as observe how they work,” Reeves says.
Central to the microscope are advances made by Professor
of Chemistry, Biochemisty and Molecular Biology Akos Vertes,
who uses lasers and mass spectrometry to gather and identify
individual proteins. In his research, tissue is exposed to
a 3-micron infrared laser, exciting the water molecules around
proteins, which conveniently “pop out” of the
cell intact. This technique previously had to be done in a
vacuum, but Vertes developed a method to do it in normal ambient
conditions, greatly facilitating the technique.
The extracted proteins are injected into a mass spectrometer
to be analyzed and identified. This information is then used
by the protein microscope to detect wether a particular protein
is present in tissue samples.
“The protein microscope uses an optical fiber that
has been sharpened to a fine point so that the microscope’s
laser light can focus on a spot 100 or 200 times smaller than
is possible with conventional microscopes,” Reeves explains.
“Our laser spot is only about two-tenths of a micron
in diameter, which is less than the width of a single cell.”
“We can see individual features on a single cell and
detect which proteins are there. We can also see how their
population changes as the tissue is stimulated with various
materials or actions. We can eventually develop an in-depth
understanding of proteins, and this will help us develop models
to be able to mathematically predict their roles.”
Testing and implementing the protein microscope will depend
upon the knowledge of well-studied cellular systems. One such
exemplary system is being provided by Fatah Kashanchi, professor
of biochemistry and molecular biology and a specialist in
the proteomics of HIV and leukemia viruses. Kashanchi is using
mass spectroscopy to determine the proteins present in the
membranes of cells infected with the HIV virus, which will
be later studied with the microscope system. (See article
on GW’s HIV/AIDS research on Page 11.)
The project’s first area of research focuses on a narrow,
one-tenth of a micron region where the nerve meets the muscles,
called the neuromuscular junctions. This builds upon the research
of Eric Hoffman, professor of pediatrics, biochemistry, and
molecular biology. The goal is to determine which proteins
are active in healthy or diseased subjects. The findings could
further Hoffman’s work to understand the causes of childhood
ALS (Lou Gehrig’s disease), which can be caused by genetic
abnormalities or by protein malfunctions.
Hoffman’s lab group has just completed the first paper
from the protein microscope project. The work investigates
the proteomic and DNA analysis of a torpedo fish organ that
has a mass of neuromuscular junctions.
Speaking to the microscope’s practical applications,
Reeves says, “If we can understand the role of proteins
at the molecular level, it could lead to development of drugs
that could be used to block a protein in overabundance or
stimulate production of a protein that’s lacking. Through
the protein microscope project, we are looking for the information
we need to develop these protein-based cures.”
Motion Capture And Analysis Laboratory (MOCA)
David Chichka: Assistant Professor,
Engineering and Applied Science
Jerome Danoff: Associate Professor, Exercise
Science
Kenneth Fine: Assistant Professor,
Orthopedic Surgery
James Hahn: Professor, Engineering
and Applied Science
Michael Harris-Love: Assistant Professor,
Health Care Sciences
Kerr-Jai Lu: Assistant Professor,
Engineering and Applied Science
James Michelson: Professor, Orthopaedic
Surgery
John Philbeck: Associate Professor,
Psychology
Margaret M. Plack: Associate Professor,
Physical Therapy
Brian G. Richmond: Associate Professor,
Anthropology
Maida Withers: Professor, Dance
Athletes, physical therapy patients, and dancers are among
many groups who could potentially benefit from one of GW’s
most diverse interdisciplinary research efforts: the Motion
Capture and Analysis Laboratory (MOCA). The laboratory is
a joint effort involving eight investigators from eight departments,
as well as GW’s Institute for Biomedical Engineering
and Institute for Computer Graphics.
“The lab became operational in November 2005 with the
help of the Research Enhancement Fund. It has a Vicon optical
system and computing equipment that can capture the motion
of just about anything,” explains James Hahn, professor
of engineering and applied science, chair of the Department
of Computer Science, and director for both institutes. “Reflective
markers are attached to the subject being studied, and six
infrared cameras track the reflectors’ motion, triangulate
their positions, and record the motion in four dimensions:
x, y, z, and time. This motion data is then visualized and
analyzed using computer graphics.
“Once motion is captured, we can visualize it from
any vantage point,” Hahn says. “We can analyze
the data to calculate and visualize many types of useful information.
And we can use inverse dynamics to calculate the physical
properties of the motion, such as impacts on the body.”

This is a motion-capture image of dancer Wendell Cooper
created in GW’s MOCA.
Can Kirmizibayrak
|

|
One application for MOCA comes from a growing area of medical
research: the assessment of movement dysfunction, which afflicts
patients with disorders such as Parkinson’s disease,
multiple sclerosis, and arthritis. Movement dysfunction usually
is characterized by subjective observations in clinical and
research settings. Michael Harris-Love, assistant professor
of health care sciences, will be using MOCA to capture reaching
performance and help develop force-control testing as an objective
measure of movement dysfunction.
Another medical research project is using MOCA to help determine
the accuracy of optical motion-capture systems used in image-guided
surgery. The research is part of a $2.8 million project funded
by the National Institutes of Health involving GW’s
School of Engineering and Applied Science and School of Medicine
and Health Sciences.
“We are working on an image-based approach using computer-vision
techniques to register pre-operative CT to intra-operative
patients,” Hahn says. “We are using MOCA to analyze
the infrared motion-capture systems and compare them to a
more-sophisticated approach we are developing.”
MOCA also is being used to further research on athletics,
be it to improve performance or prevent injuries. Kenneth
Fine, assistant professor of orthopaedic surgery and GW varsity
team physician, is studying the motions of soccer players.
And Jerome Danoff, associate professor of exercise science,
is analyzing Chi running, which is claimed to be more efficient,
less tiring, and less injury provoking than standard running.
The analysis and visualization of athletes represent an extension
of an ongoing project by the group with USA Swimming, the
governing body for U.S. Olympic swimmers.
Using MOCA helped researchers better understand how people
keep track of their location during the complex body motions
involved in walking. This research, led by John Philbeck,
associate professor of psychology, is important in understanding
fundamental issues behind human locomotion and navigation.
MOCA also was used by Kerr-Jai Lu, assistant professor of
engineering and applied science, to help develop a biomimetic
fin for an unmanned underwater vehicle developed in the Naval
Research Laboratory. The biomimetic fin—inspired by
the pectoral fin of a wrasse fish—is designed to mimic
the natural fin motion. Using MOCA, the kinematics and shape
change of the fin prototype are precisely captured. This information
is entered into a computational fluid dynamics code to estimate
the fin’s force production capability, which, in turn,
helps improve the overall vehicle design.
Brian Richmond, associate professor of anthropology, is using
MOCA to study fossil foot bones and trace the origin of the
uniquely human “toe-off” during gait. He is using
MOCA to precisely measure movements of the toes and forefoot
during walking and running to clarify the relationship between
the function and structure of the foot skeleton.
MOCA is being used by Maida Withers, professor of dance,
for an interactive dance performance. The ultimate objective
is to capture the dancers’ motion, create a visualization
in the form of computer animations, and project it onto the
stage where the real dancer is performing. Thus the virtual
image and the real dancer interact to create a multidimensional
dance work.
“In MOCA, we have investigators from many different
disciplines working together who would normally not interact,”
Hahn says. “The glue that holds everything together
is the ability to take something as fleeting as motion and
manipulate it in the computer. We are using this tool to look
at the world through the viewpoint of a wide range of disciplines.”
Kathleen Kocks is a freelance writer located in New Orleans.
|